skimage.color#
Color space conversion.
Stain to RGB color space conversion. |
|
Convert an image array to a new color space. |
|
Euclidean distance between two points in Lab color space |
|
Color difference as given by the CIEDE 2000 standard. |
|
Color difference according to CIEDE 94 standard |
|
Color difference from the CMC l:c standard. |
|
Create an RGB representation of a gray-level image. |
|
Create a RGBA representation of a gray-level image. |
|
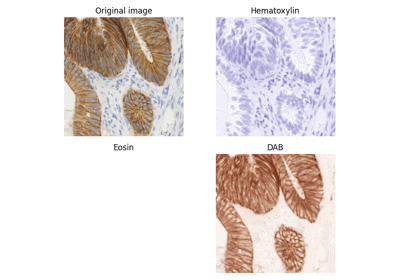
Haematoxylin-Eosin-DAB (HED) to RGB color space conversion. |
|
HSV to RGB color space conversion. |
|
Convert image in CIE-LAB to CIE-LCh color space. |
|
Convert image in CIE-LAB to sRGB color space. |
|
Convert image in CIE-LAB to XYZ color space. |
|
Return an RGB image where color-coded labels are painted over the image. |
|
Convert image in CIE-LCh to CIE-LAB color space. |
|
Luv to RGB color space conversion. |
|
CIE-Luv to XYZ color space conversion. |
|
Compute luminance of an RGB image. |
|
RGB to Haematoxylin-Eosin-DAB (HED) color space conversion. |
|
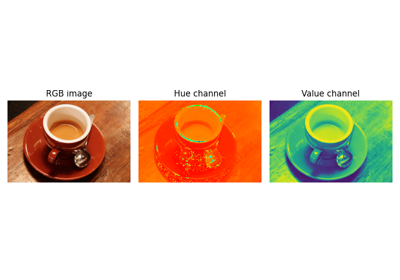
RGB to HSV color space conversion. |
|
Conversion from the sRGB color space (IEC 61966-2-1:1999) to the CIE Lab colorspace under the given illuminant and observer. |
|
RGB to CIE-Luv color space conversion. |
|
RGB to RGB CIE color space conversion. |
|
RGB to XYZ color space conversion. |
|
RGB to YCbCr color space conversion. |
|
RGB to YDbDr color space conversion. |
|
RGB to YIQ color space conversion. |
|
RGB to YPbPr color space conversion. |
|
RGB to YUV color space conversion. |
|
RGBA to RGB conversion using alpha blending [1]. |
|
RGB CIE to RGB color space conversion. |
|
RGB to stain color space conversion. |
|
XYZ to CIE-LAB color space conversion. |
|
XYZ to CIE-Luv color space conversion. |
|
XYZ to RGB color space conversion. |
|
Get the CIE XYZ tristimulus values. |
|
YCbCr to RGB color space conversion. |
|
YDbDr to RGB color space conversion. |
|
YIQ to RGB color space conversion. |
|
YPbPr to RGB color space conversion. |
|
YUV to RGB color space conversion. |
- skimage.color.combine_stains(stains, conv_matrix, *, channel_axis=-1)[source]#
Stain to RGB color space conversion.
- Parameters:
- stains(…, C=3, …) array_like
The image in stain color space. By default, the final dimension denotes channels.
- conv_matrixndarray
The stain separation matrix as described by G. Landini [1].
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
stainsis not at least 2-D with shape (…, C=3, …).
Notes
Stain combination matrices available in the
colormodule and their respective colorspace:rgb_from_hed: Hematoxylin + Eosin + DABrgb_from_hdx: Hematoxylin + DABrgb_from_fgx: Feulgen + Light Greenrgb_from_bex: Giemsa stain : Methyl Blue + Eosinrgb_from_rbd: FastRed + FastBlue + DABrgb_from_gdx: Methyl Green + DABrgb_from_hax: Hematoxylin + AECrgb_from_bro: Blue matrix Anilline Blue + Red matrix Azocarmine + Orange matrix Orange-Grgb_from_bpx: Methyl Blue + Ponceau Fuchsinrgb_from_ahx: Alcian Blue + Hematoxylinrgb_from_hpx: Hematoxylin + PAS
References
[1][2]A. C. Ruifrok and D. A. Johnston, “Quantification of histochemical staining by color deconvolution,” Anal. Quant. Cytol. Histol., vol. 23, no. 4, pp. 291–299, Aug. 2001.
Examples
>>> from skimage import data >>> from skimage.color import (separate_stains, combine_stains, ... hdx_from_rgb, rgb_from_hdx) >>> ihc = data.immunohistochemistry() >>> ihc_hdx = separate_stains(ihc, hdx_from_rgb) >>> ihc_rgb = combine_stains(ihc_hdx, rgb_from_hdx)
- skimage.color.convert_colorspace(arr, fromspace, tospace, *, channel_axis=-1)[source]#
Convert an image array to a new color space.
- Valid color spaces are:
‘RGB’, ‘HSV’, ‘RGB CIE’, ‘XYZ’, ‘YUV’, ‘YIQ’, ‘YPbPr’, ‘YCbCr’, ‘YDbDr’
- Parameters:
- arr(…, C=3, …) array_like
The image to convert. By default, the final dimension denotes channels.
- fromspacestr
The color space to convert from. Can be specified in lower case.
- tospacestr
The color space to convert to. Can be specified in lower case.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The converted image. Same dimensions as input.
- Raises:
- ValueError
If fromspace is not a valid color space
- ValueError
If tospace is not a valid color space
Notes
Conversion is performed through the “central” RGB color space, i.e. conversion from XYZ to HSV is implemented as
XYZ -> RGB -> HSVinstead of directly.Examples
>>> from skimage import data >>> img = data.astronaut() >>> img_hsv = convert_colorspace(img, 'RGB', 'HSV')
- skimage.color.deltaE_cie76(lab1, lab2, channel_axis=-1)[source]#
Euclidean distance between two points in Lab color space
- Parameters:
- lab1array_like
reference color (Lab colorspace)
- lab2array_like
comparison color (Lab colorspace)
- channel_axisint, optional
This parameter indicates which axis of the arrays corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- dEarray_like
distance between colors
lab1andlab2
References
[2]A. R. Robertson, “The CIE 1976 color-difference formulae,” Color Res. Appl. 2, 7-11 (1977).
- skimage.color.deltaE_ciede2000(lab1, lab2, kL=1, kC=1, kH=1, *, channel_axis=-1)[source]#
Color difference as given by the CIEDE 2000 standard.
CIEDE 2000 is a major revision of CIDE94. The perceptual calibration is largely based on experience with automotive paint on smooth surfaces.
- Parameters:
- lab1array_like
reference color (Lab colorspace)
- lab2array_like
comparison color (Lab colorspace)
- kLfloat (range), optional
lightness scale factor, 1 for “acceptably close”; 2 for “imperceptible” see deltaE_cmc
- kCfloat (range), optional
chroma scale factor, usually 1
- kHfloat (range), optional
hue scale factor, usually 1
- channel_axisint, optional
This parameter indicates which axis of the arrays corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- deltaEarray_like
The distance between
lab1andlab2
Notes
CIEDE 2000 assumes parametric weighting factors for the lightness, chroma, and hue (
kL,kC,kHrespectively). These default to 1.References
[3]M. Melgosa, J. Quesada, and E. Hita, “Uniformity of some recent color metrics tested with an accurate color-difference tolerance dataset,” Appl. Opt. 33, 8069-8077 (1994).
- skimage.color.deltaE_ciede94(lab1, lab2, kH=1, kC=1, kL=1, k1=0.045, k2=0.015, *, channel_axis=-1)[source]#
Color difference according to CIEDE 94 standard
Accommodates perceptual non-uniformities through the use of application specific scale factors (
kH,kC,kL,k1, andk2).- Parameters:
- lab1array_like
reference color (Lab colorspace)
- lab2array_like
comparison color (Lab colorspace)
- kHfloat, optional
Hue scale
- kCfloat, optional
Chroma scale
- kLfloat, optional
Lightness scale
- k1float, optional
first scale parameter
- k2float, optional
second scale parameter
- channel_axisint, optional
This parameter indicates which axis of the arrays corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- dEarray_like
color difference between
lab1andlab2
Notes
deltaE_ciede94 is not symmetric with respect to lab1 and lab2. CIEDE94 defines the scales for the lightness, hue, and chroma in terms of the first color. Consequently, the first color should be regarded as the “reference” color.
kL,k1,k2depend on the application and default to the values suggested for graphic artsParameter
Graphic Arts
Textiles
kL1.000
2.000
k10.045
0.048
k20.015
0.014
References
- skimage.color.deltaE_cmc(lab1, lab2, kL=1, kC=1, *, channel_axis=-1)[source]#
Color difference from the CMC l:c standard.
This color difference was developed by the Colour Measurement Committee (CMC) of the Society of Dyers and Colourists (United Kingdom). It is intended for use in the textile industry.
The scale factors
kL,kCset the weight given to differences in lightness and chroma relative to differences in hue. The usual values arekL=2,kC=1for “acceptability” andkL=1,kC=1for “imperceptibility”. Colors withdE > 1are “different” for the given scale factors.- Parameters:
- lab1array_like
reference color (Lab colorspace)
- lab2array_like
comparison color (Lab colorspace)
- channel_axisint, optional
This parameter indicates which axis of the arrays corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- dEarray_like
distance between colors
lab1andlab2
Notes
deltaE_cmc the defines the scales for the lightness, hue, and chroma in terms of the first color. Consequently
deltaE_cmc(lab1, lab2) != deltaE_cmc(lab2, lab1)References
[3]F. J. J. Clarke, R. McDonald, and B. Rigg, “Modification to the JPC79 colour-difference formula,” J. Soc. Dyers Colour. 100, 128-132 (1984).
- skimage.color.gray2rgb(image, *, channel_axis=-1)[source]#
Create an RGB representation of a gray-level image.
- Parameters:
- imagearray_like
Input image.
- channel_axisint, optional
This parameter indicates which axis of the output array will correspond to channels.
- Returns:
- rgb(…, C=3, …) ndarray
RGB image. A new dimension of length 3 is added to input image.
Notes
If the input is a 1-dimensional image of shape
(M,), the output will be shape(M, C=3).
Region Boundary based Region adjacency graphs (RAGs)
Region Boundary based Region adjacency graphs (RAGs)
- skimage.color.gray2rgba(image, alpha=None, *, channel_axis=-1)[source]#
Create a RGBA representation of a gray-level image.
- Parameters:
- imagearray_like
Input image.
- alphaarray_like, optional
Alpha channel of the output image. It may be a scalar or an array that can be broadcast to
image. If not specified it is set to the maximum limit corresponding to theimagedtype.- channel_axisint, optional
This parameter indicates which axis of the output array will correspond to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- rgbandarray
RGBA image. A new dimension of length 4 is added to input image shape.
- skimage.color.hed2rgb(hed, *, channel_axis=-1)[source]#
Haematoxylin-Eosin-DAB (HED) to RGB color space conversion.
- Parameters:
- hed(…, C=3, …) array_like
The image in the HED color space. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB. Same dimensions as input.
- Raises:
- ValueError
If
hedis not at least 2-D with shape (…, C=3, …).
References
[1]A. C. Ruifrok and D. A. Johnston, “Quantification of histochemical staining by color deconvolution.,” Analytical and quantitative cytology and histology / the International Academy of Cytology [and] American Society of Cytology, vol. 23, no. 4, pp. 291-9, Aug. 2001.
Examples
>>> from skimage import data >>> from skimage.color import rgb2hed, hed2rgb >>> ihc = data.immunohistochemistry() >>> ihc_hed = rgb2hed(ihc) >>> ihc_rgb = hed2rgb(ihc_hed)
- skimage.color.hsv2rgb(hsv, *, channel_axis=-1)[source]#
HSV to RGB color space conversion.
- Parameters:
- hsv(…, C=3, …) array_like
The image in HSV format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
hsvis not at least 2-D with shape (…, C=3, …).
Notes
Conversion between RGB and HSV color spaces results in some loss of precision, due to integer arithmetic and rounding [1].
References
Examples
>>> from skimage import data >>> img = data.astronaut() >>> img_hsv = rgb2hsv(img) >>> img_rgb = hsv2rgb(img_hsv)
- skimage.color.lab2lch(lab, *, channel_axis=-1)[source]#
Convert image in CIE-LAB to CIE-LCh color space.
CIE-LCh is the cylindrical representation of the CIE-LAB (Cartesian) color space.
- Parameters:
- lab(…, C=3, …) array_like
The input image in CIE-LAB color space. Unless
channel_axisis set, the final dimension denotes the CIE-LAB channels. The L* values range from 0 to 100; the a* and b* values range from -128 to 127.- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in CIE-LCh color space, of same shape as input.
- Raises:
- ValueError
If
labdoes not have at least 3 channels (i.e., L*, a*, and b*).
See also
Notes
The h channel (i.e., hue) is expressed as an angle in range
(0, 2*pi).References
Examples
>>> from skimage import data >>> from skimage.color import rgb2lab, lab2lch >>> img = data.astronaut() >>> img_lab = rgb2lab(img) >>> img_lch = lab2lch(img_lab)
- skimage.color.lab2rgb(lab, illuminant='D65', observer='2', *, channel_axis=-1)[source]#
Convert image in CIE-LAB to sRGB color space.
- Parameters:
- lab(…, C=3, …) array_like
The input image in CIE-LAB color space. Unless
channel_axisis set, the final dimension denotes the CIE-LAB channels. The L* values range from 0 to 100; the a* and b* values range from -128 to 127.- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}, optional
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}, optional
The aperture angle of the observer.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in sRGB color space, of same shape as input.
- Raises:
- ValueError
If
labis not at least 2-D with shape (…, C=3, …).
See also
Notes
This function uses
lab2xyz()andxyz2rgb(). The CIE XYZ tristimulus values are x_ref = 95.047, y_ref = 100., and z_ref = 108.883. See functionxyz_tristimulus_values()for a list of supported illuminants.References
- skimage.color.lab2xyz(lab, illuminant='D65', observer='2', *, channel_axis=-1)[source]#
Convert image in CIE-LAB to XYZ color space.
- Parameters:
- lab(…, C=3, …) array_like
The input image in CIE-LAB color space. Unless
channel_axisis set, the final dimension denotes the CIE-LAB channels. The L* values range from 0 to 100; the a* and b* values range from -128 to 127.- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}, optional
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}, optional
The aperture angle of the observer.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in XYZ color space, of same shape as input.
- Raises:
- ValueError
If
labis not at least 2-D with shape (…, C=3, …).- ValueError
If either the illuminant or the observer angle are not supported or unknown.
- UserWarning
If any of the pixels are invalid (Z < 0).
See also
Notes
The CIE XYZ tristimulus values are x_ref = 95.047, y_ref = 100., and z_ref = 108.883. See function
xyz_tristimulus_values()for a list of supported illuminants.References
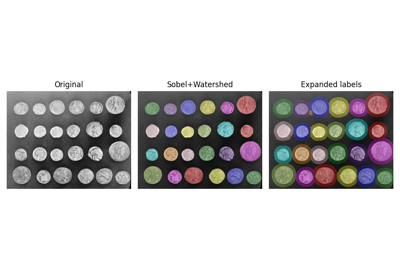
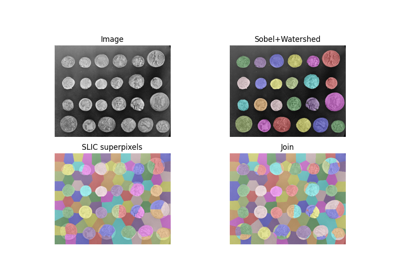
- skimage.color.label2rgb(label, image=None, colors=None, alpha=0.3, bg_label=0, bg_color=(0, 0, 0), image_alpha=1, kind='overlay', *, saturation=0, channel_axis=-1)[source]#
Return an RGB image where color-coded labels are painted over the image.
- Parameters:
- labelndarray
Integer array of labels with the same shape as
image.- imagendarray, optional
Image used as underlay for labels. It should have the same shape as
labels, optionally with an additional RGB (channels) axis. Ifimageis an RGB image, it is converted to grayscale before coloring.- colorslist, optional
List of colors. If the number of labels exceeds the number of colors, then the colors are cycled.
- alphafloat [0, 1], optional
Opacity of colorized labels. Ignored if image is
None.- bg_labelint, optional
Label that’s treated as the background. If
bg_labelis specified,bg_colorisNone, andkindisoverlay, background is not painted by any colors.- bg_colorstr or array, optional
Background color. Must be a name in
skimage.color.color_dictor RGB float values between [0, 1].- image_alphafloat [0, 1], optional
Opacity of the image.
- kindstring, one of {‘overlay’, ‘avg’}
The kind of color image desired. ‘overlay’ cycles over defined colors and overlays the colored labels over the original image. ‘avg’ replaces each labeled segment with its average color, for a stained-class or pastel painting appearance.
- saturationfloat [0, 1], optional
Parameter to control the saturation applied to the original image between fully saturated (original RGB,
saturation=1) and fully unsaturated (grayscale,saturation=0). Only applies whenkind='overlay'.- channel_axisint, optional
This parameter indicates which axis of the output array will correspond to channels. If
imageis provided, this must also match the axis ofimagethat corresponds to channels.Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- resultndarray of float, same shape as
image The result of blending a cycling colormap (
colors) for each distinct value inlabelwith the image, at a certain alpha value.
- resultndarray of float, same shape as
Comparing edge-based and region-based segmentation
Comparing edge-based and region-based segmentation
Use pixel graphs to find an object’s geodesic center
Use pixel graphs to find an object's geodesic center
- skimage.color.lch2lab(lch, *, channel_axis=-1)[source]#
Convert image in CIE-LCh to CIE-LAB color space.
CIE-LCh is the cylindrical representation of the CIE-LAB (Cartesian) color space.
- Parameters:
- lch(…, C=3, …) array_like
The input image in CIE-LCh color space. Unless
channel_axisis set, the final dimension denotes the CIE-LAB channels. The L* values range from 0 to 100; the C values range from 0 to 100; the h values range from 0 to2*pi.- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in CIE-LAB format, of same shape as input.
- Raises:
- ValueError
If
lchdoes not have at least 3 channels (i.e., L*, C, and h).
See also
Notes
The h channel (i.e., hue) is expressed as an angle in range
(0, 2*pi).References
Examples
>>> from skimage import data >>> from skimage.color import rgb2lab, lch2lab, lab2lch >>> img = data.astronaut() >>> img_lab = rgb2lab(img) >>> img_lch = lab2lch(img_lab) >>> img_lab2 = lch2lab(img_lch)
- skimage.color.luv2rgb(luv, *, channel_axis=-1)[source]#
Luv to RGB color space conversion.
- Parameters:
- luv(…, C=3, …) array_like
The image in CIE Luv format. By default, the final dimension denotes channels.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
luvis not at least 2-D with shape (…, C=3, …).
Notes
This function uses luv2xyz and xyz2rgb.
- skimage.color.luv2xyz(luv, illuminant='D65', observer='2', *, channel_axis=-1)[source]#
CIE-Luv to XYZ color space conversion.
- Parameters:
- luv(…, C=3, …) array_like
The image in CIE-Luv format. By default, the final dimension denotes channels.
- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}, optional
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}, optional
The aperture angle of the observer.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in XYZ format. Same dimensions as input.
- Raises:
- ValueError
If
luvis not at least 2-D with shape (…, C=3, …).- ValueError
If either the illuminant or the observer angle are not supported or unknown.
Notes
XYZ conversion weights use observer=2A. Reference whitepoint for D65 Illuminant, with XYZ tristimulus values of
(95.047, 100., 108.883). See functionxyz_tristimulus_values()for a list of supported illuminants.References
- skimage.color.rgb2gray(rgb, *, channel_axis=-1)[source]#
Compute luminance of an RGB image.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- Returns:
- outndarray
The luminance image - an array which is the same size as the input array, but with the channel dimension removed.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
The weights used in this conversion are calibrated for contemporary CRT phosphors:
Y = 0.2125 R + 0.7154 G + 0.0721 B
If there is an alpha channel present, it is ignored.
References
Examples
>>> from skimage.color import rgb2gray >>> from skimage import data >>> img = data.astronaut() >>> img_gray = rgb2gray(img)

Use pixel graphs to find an object’s geodesic center
Use pixel graphs to find an object's geodesic center
Gabors / Primary Visual Cortex “Simple Cells” from an Image
Gabors / Primary Visual Cortex "Simple Cells" from an Image
Full tutorial on calibrating Denoisers Using J-Invariance
Full tutorial on calibrating Denoisers Using J-Invariance
Removing small objects in grayscale images with a top hat filter
Removing small objects in grayscale images with a top hat filter
Using Polar and Log-Polar Transformations for Registration
Using Polar and Log-Polar Transformations for Registration
Region Boundary based Region adjacency graphs (RAGs)
Region Boundary based Region adjacency graphs (RAGs)
Comparison of segmentation and superpixel algorithms
Comparison of segmentation and superpixel algorithms
- skimage.color.rgb2hed(rgb, *, channel_axis=-1)[source]#
RGB to Haematoxylin-Eosin-DAB (HED) color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in HED format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
References
[1]A. C. Ruifrok and D. A. Johnston, “Quantification of histochemical staining by color deconvolution.,” Analytical and quantitative cytology and histology / the International Academy of Cytology [and] American Society of Cytology, vol. 23, no. 4, pp. 291-9, Aug. 2001.
Examples
>>> from skimage import data >>> from skimage.color import rgb2hed >>> ihc = data.immunohistochemistry() >>> ihc_hed = rgb2hed(ihc)
- skimage.color.rgb2hsv(rgb, *, channel_axis=-1)[source]#
RGB to HSV color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in HSV format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
Conversion between RGB and HSV color spaces results in some loss of precision, due to integer arithmetic and rounding [1].
References
Examples
>>> from skimage import color >>> from skimage import data >>> img = data.astronaut() >>> img_hsv = color.rgb2hsv(img)
- skimage.color.rgb2lab(rgb, illuminant='D65', observer='2', *, channel_axis=-1)[source]#
Conversion from the sRGB color space (IEC 61966-2-1:1999) to the CIE Lab colorspace under the given illuminant and observer.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}, optional
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}, optional
The aperture angle of the observer.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in Lab format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
RGB is a device-dependent color space so, if you use this function, be sure that the image you are analyzing has been mapped to the sRGB color space.
This function uses rgb2xyz and xyz2lab. By default Observer=”2”, Illuminant=”D65”. CIE XYZ tristimulus values x_ref=95.047, y_ref=100., z_ref=108.883. See function
xyz_tristimulus_values()for a list of supported illuminants.References
- skimage.color.rgb2luv(rgb, *, channel_axis=-1)[source]#
RGB to CIE-Luv color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in CIE Luv format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
This function uses rgb2xyz and xyz2luv.
References
- skimage.color.rgb2rgbcie(rgb, *, channel_axis=-1)[source]#
RGB to RGB CIE color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB CIE format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
References
Examples
>>> from skimage import data >>> from skimage.color import rgb2rgbcie >>> img = data.astronaut() >>> img_rgbcie = rgb2rgbcie(img)
- skimage.color.rgb2xyz(rgb, *, channel_axis=-1)[source]#
RGB to XYZ color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in XYZ format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
The CIE XYZ color space is derived from the CIE RGB color space. Note however that this function converts from sRGB.
References
Examples
>>> from skimage import data >>> img = data.astronaut() >>> img_xyz = rgb2xyz(img)
- skimage.color.rgb2ycbcr(rgb, *, channel_axis=-1)[source]#
RGB to YCbCr color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in YCbCr format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
Y is between 16 and 235. This is the color space commonly used by video codecs; it is sometimes incorrectly called “YUV”.
References
- skimage.color.rgb2ydbdr(rgb, *, channel_axis=-1)[source]#
RGB to YDbDr color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in YDbDr format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
This is the color space commonly used by video codecs. It is also the reversible color transform in JPEG2000.
References
- skimage.color.rgb2yiq(rgb, *, channel_axis=-1)[source]#
RGB to YIQ color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in YIQ format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
- skimage.color.rgb2ypbpr(rgb, *, channel_axis=-1)[source]#
RGB to YPbPr color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in YPbPr format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
References
- skimage.color.rgb2yuv(rgb, *, channel_axis=-1)[source]#
RGB to YUV color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in YUV format. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
Y is between 0 and 1. Use YCbCr instead of YUV for the color space commonly used by video codecs, where Y ranges from 16 to 235.
References
- skimage.color.rgba2rgb(rgba, background=(1, 1, 1), *, channel_axis=-1)[source]#
RGBA to RGB conversion using alpha blending [1].
- Parameters:
- rgba(…, C=4, …) array_like
The image in RGBA format. By default, the final dimension denotes channels.
- backgroundarray_like
The color of the background to blend the image with (3 floats between 0 to 1 - the RGB value of the background).
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
rgbais not at least 2D with shape (…, 4, …).
References
Examples
>>> from skimage import color >>> from skimage import data >>> img_rgba = data.logo() >>> img_rgb = color.rgba2rgb(img_rgba)
- skimage.color.rgbcie2rgb(rgbcie, *, channel_axis=-1)[source]#
RGB CIE to RGB color space conversion.
- Parameters:
- rgbcie(…, C=3, …) array_like
The image in RGB CIE format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
rgbcieis not at least 2-D with shape (…, C=3, …).
References
Examples
>>> from skimage import data >>> from skimage.color import rgb2rgbcie, rgbcie2rgb >>> img = data.astronaut() >>> img_rgbcie = rgb2rgbcie(img) >>> img_rgb = rgbcie2rgb(img_rgbcie)
- skimage.color.separate_stains(rgb, conv_matrix, *, channel_axis=-1)[source]#
RGB to stain color space conversion.
- Parameters:
- rgb(…, C=3, …) array_like
The image in RGB format. By default, the final dimension denotes channels.
- conv_matrixndarray
The stain separation matrix as described by G. Landini [1].
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in stain color space. Same dimensions as input.
- Raises:
- ValueError
If
rgbis not at least 2-D with shape (…, C=3, …).
Notes
Stain separation matrices available in the
colormodule and their respective colorspace:hed_from_rgb: Hematoxylin + Eosin + DABhdx_from_rgb: Hematoxylin + DABfgx_from_rgb: Feulgen + Light Greenbex_from_rgb: Giemsa stain : Methyl Blue + Eosinrbd_from_rgb: FastRed + FastBlue + DABgdx_from_rgb: Methyl Green + DABhax_from_rgb: Hematoxylin + AECbro_from_rgb: Blue matrix Anilline Blue + Red matrix Azocarmine + Orange matrix Orange-Gbpx_from_rgb: Methyl Blue + Ponceau Fuchsinahx_from_rgb: Alcian Blue + Hematoxylinhpx_from_rgb: Hematoxylin + PAS
This implementation borrows some ideas from DIPlib [2], e.g. the compensation using a small value to avoid log artifacts when calculating the Beer-Lambert law.
References
[1][3]A. C. Ruifrok and D. A. Johnston, “Quantification of histochemical staining by color deconvolution,” Anal. Quant. Cytol. Histol., vol. 23, no. 4, pp. 291–299, Aug. 2001.
Examples
>>> from skimage import data >>> from skimage.color import separate_stains, hdx_from_rgb >>> ihc = data.immunohistochemistry() >>> ihc_hdx = separate_stains(ihc, hdx_from_rgb)
- skimage.color.xyz2lab(xyz, illuminant='D65', observer='2', *, channel_axis=-1)[source]#
XYZ to CIE-LAB color space conversion.
- Parameters:
- xyz(…, C=3, …) array_like
The image in XYZ format. By default, the final dimension denotes channels.
- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}, optional
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}, optional
One of: 2-degree observer, 10-degree observer, or ‘R’ observer as in R function grDevices::convertColor.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in CIE-LAB format. Same dimensions as input.
- Raises:
- ValueError
If
xyzis not at least 2-D with shape (…, C=3, …).- ValueError
If either the illuminant or the observer angle is unsupported or unknown.
Notes
By default Observer=”2”, Illuminant=”D65”. CIE XYZ tristimulus values x_ref=95.047, y_ref=100., z_ref=108.883. See function
xyz_tristimulus_values()for a list of supported illuminants.References
Examples
>>> from skimage import data >>> from skimage.color import rgb2xyz, xyz2lab >>> img = data.astronaut() >>> img_xyz = rgb2xyz(img) >>> img_lab = xyz2lab(img_xyz)
- skimage.color.xyz2luv(xyz, illuminant='D65', observer='2', *, channel_axis=-1)[source]#
XYZ to CIE-Luv color space conversion.
- Parameters:
- xyz(…, C=3, …) array_like
The image in XYZ format. By default, the final dimension denotes channels.
- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}, optional
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}, optional
The aperture angle of the observer.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in CIE-Luv format. Same dimensions as input.
- Raises:
- ValueError
If
xyzis not at least 2-D with shape (…, C=3, …).- ValueError
If either the illuminant or the observer angle are not supported or unknown.
Notes
By default XYZ conversion weights use observer=2A. Reference whitepoint for D65 Illuminant, with XYZ tristimulus values of
(95.047, 100., 108.883). See functionxyz_tristimulus_values()for a list of supported illuminants.References
Examples
>>> from skimage import data >>> from skimage.color import rgb2xyz, xyz2luv >>> img = data.astronaut() >>> img_xyz = rgb2xyz(img) >>> img_luv = xyz2luv(img_xyz)
- skimage.color.xyz2rgb(xyz, *, channel_axis=-1)[source]#
XYZ to RGB color space conversion.
- Parameters:
- xyz(…, C=3, …) array_like
The image in XYZ format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
xyzis not at least 2-D with shape (…, C=3, …).
Notes
The CIE XYZ color space is derived from the CIE RGB color space. Note however that this function converts to sRGB.
References
Examples
>>> from skimage import data >>> from skimage.color import rgb2xyz, xyz2rgb >>> img = data.astronaut() >>> img_xyz = rgb2xyz(img) >>> img_rgb = xyz2rgb(img_xyz)
- skimage.color.xyz_tristimulus_values(*, illuminant, observer, dtype=<class 'float'>)[source]#
Get the CIE XYZ tristimulus values.
Given an illuminant and observer, this function returns the CIE XYZ tristimulus values [2] scaled such that \(Y = 1\).
- Parameters:
- illuminant{“A”, “B”, “C”, “D50”, “D55”, “D65”, “D75”, “E”}
The name of the illuminant (the function is NOT case sensitive).
- observer{“2”, “10”, “R”}
One of: 2-degree observer, 10-degree observer, or ‘R’ observer as in R function
grDevices::convertColor[3].- dtypedtype, optional
Output data type.
- Returns:
- valuesarray
Array with 3 elements \(X, Y, Z\) containing the CIE XYZ tristimulus values of the given illuminant.
- Raises:
- ValueError
If either the illuminant or the observer angle are not supported or unknown.
Notes
The CIE XYZ tristimulus values are calculated from \(x, y\) [1], using the formula
\[X = x / y\]\[Y = 1\]\[Z = (1 - x - y) / y\]The only exception is the illuminant “D65” with aperture angle 2° for backward-compatibility reasons.
References
Examples
Get the CIE XYZ tristimulus values for a “D65” illuminant for a 10 degree field of view
>>> xyz_tristimulus_values(illuminant="D65", observer="10") array([0.94809668, 1. , 1.07305136])
- skimage.color.ycbcr2rgb(ycbcr, *, channel_axis=-1)[source]#
YCbCr to RGB color space conversion.
- Parameters:
- ycbcr(…, C=3, …) array_like
The image in YCbCr format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
ycbcris not at least 2-D with shape (…, C=3, …).
Notes
Y is between 16 and 235. This is the color space commonly used by video codecs; it is sometimes incorrectly called “YUV”.
References
- skimage.color.ydbdr2rgb(ydbdr, *, channel_axis=-1)[source]#
YDbDr to RGB color space conversion.
- Parameters:
- ydbdr(…, C=3, …) array_like
The image in YDbDr format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
ydbdris not at least 2-D with shape (…, C=3, …).
Notes
This is the color space commonly used by video codecs, also called the reversible color transform in JPEG2000.
References
- skimage.color.yiq2rgb(yiq, *, channel_axis=-1)[source]#
YIQ to RGB color space conversion.
- Parameters:
- yiq(…, C=3, …) array_like
The image in YIQ format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
yiqis not at least 2-D with shape (…, C=3, …).
- skimage.color.ypbpr2rgb(ypbpr, *, channel_axis=-1)[source]#
YPbPr to RGB color space conversion.
- Parameters:
- ypbpr(…, C=3, …) array_like
The image in YPbPr format. By default, the final dimension denotes channels.
- channel_axisint, optional
This parameter indicates which axis of the array corresponds to channels.
Added in version 0.19:
channel_axiswas added in 0.19.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
ypbpris not at least 2-D with shape (…, C=3, …).
References
- skimage.color.yuv2rgb(yuv, *, channel_axis=-1)[source]#
YUV to RGB color space conversion.
- Parameters:
- yuv(…, C=3, …) array_like
The image in YUV format. By default, the final dimension denotes channels.
- Returns:
- out(…, C=3, …) ndarray
The image in RGB format. Same dimensions as input.
- Raises:
- ValueError
If
yuvis not at least 2-D with shape (…, C=3, …).
References