Note
Go to the end to download the full example code or to run this example in your browser via Binder
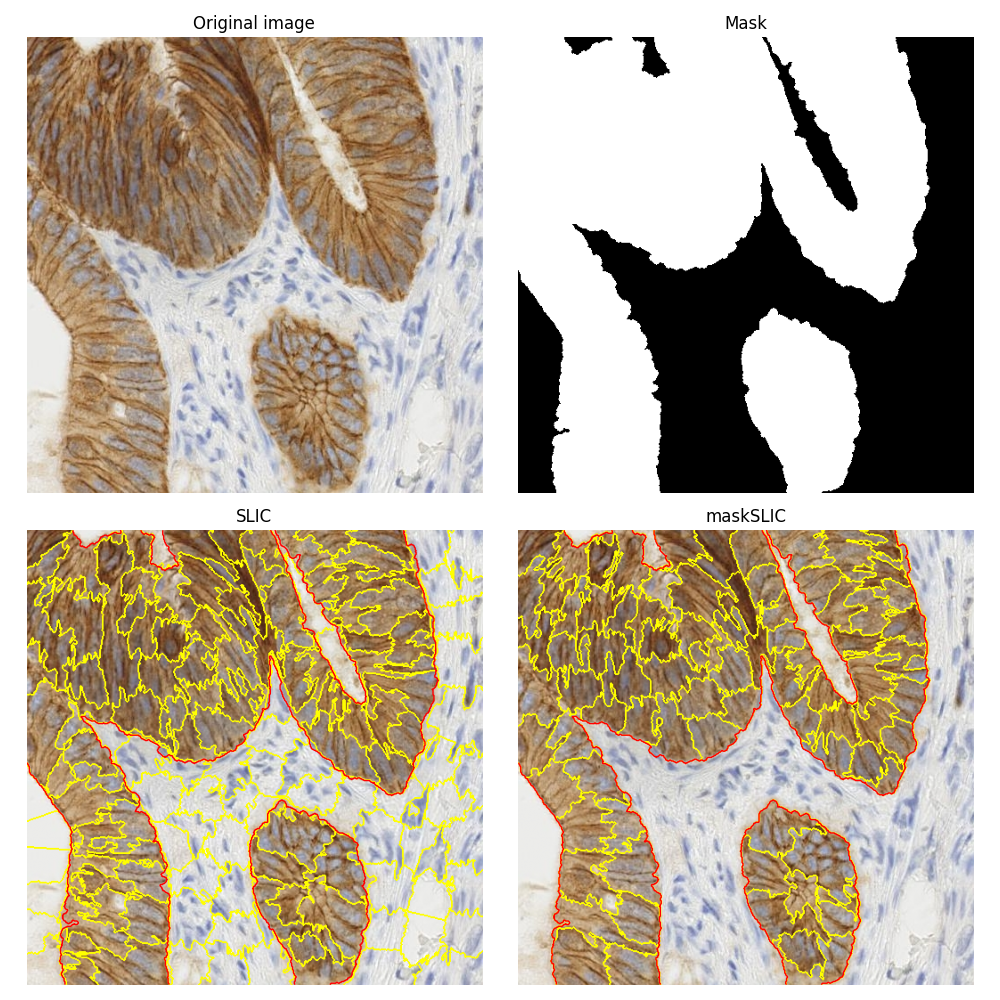
Apply maskSLIC vs SLIC#
This example is about comparing the segmentations obtained using the plain SLIC method [1] and its masked version maskSLIC [2].
To illustrate these segmentation methods, we use an image of biological tissue with immunohistochemical (IHC) staining. The same biomedical image is used in the example on how to Separate colors in immunohistochemical staining.
The maskSLIC method is an extension of the SLIC method for the generation of superpixels in a region of interest. maskSLIC is able to overcome border problems that affects SLIC method, particularely in case of irregular mask.
import matplotlib.pyplot as plt
from skimage import data
from skimage import color
from skimage import morphology
from skimage import segmentation
# Input data
img = data.immunohistochemistry()
# Compute a mask
lum = color.rgb2gray(img)
mask = morphology.remove_small_holes(
morphology.remove_small_objects(
lum < 0.7, 500),
500)
mask = morphology.opening(mask, morphology.disk(3))
# SLIC result
slic = segmentation.slic(img, n_segments=200, start_label=1)
# maskSLIC result
m_slic = segmentation.slic(img, n_segments=100, mask=mask, start_label=1)
# Display result
fig, ax_arr = plt.subplots(2, 2, sharex=True, sharey=True, figsize=(10, 10))
ax1, ax2, ax3, ax4 = ax_arr.ravel()
ax1.imshow(img)
ax1.set_title('Original image')
ax2.imshow(mask, cmap='gray')
ax2.set_title('Mask')
ax3.imshow(segmentation.mark_boundaries(img, slic))
ax3.contour(mask, colors='red', linewidths=1)
ax3.set_title('SLIC')
ax4.imshow(segmentation.mark_boundaries(img, m_slic))
ax4.contour(mask, colors='red', linewidths=1)
ax4.set_title('maskSLIC')
for ax in ax_arr.ravel():
ax.set_axis_off()
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 2.566 seconds)