Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Max-tree#
The max-tree is a hierarchical representation of an image that is the basis for a large family of morphological filters.
If we apply a threshold operation to an image, we obtain a binary image containing one or several connected components. If we apply a lower threshold, all the connected components from the higher threshold are contained in the connected components from the lower threshold. This naturally defines a hierarchy of nested components that can be represented by a tree. whenever a connected component A obtained by thresholding with threshold t1 is contained in a component B obtained by thresholding with threshold t1 < t2, we say that B is the parent of A. The resulting tree structure is called a component tree. The max-tree is a compact representation of such a component tree. [1], [2], [3], [4]
In this example we give an intuition of what a max-tree is.
References#
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
import skimage as ski
import networkx as nx
Before we start : a few helper functions
def plot_img(ax, image, title, plot_text, image_values):
"""Plot an image, overlaying image values or indices."""
ax.imshow(image, cmap='gray', aspect='equal', vmin=0, vmax=np.max(image))
ax.set_title(title)
ax.set_yticks([])
ax.set_xticks([])
for x in np.arange(-0.5, image.shape[0], 1.0):
ax.add_artist(
Line2D((x, x), (-0.5, image.shape[0] - 0.5), color='blue', linewidth=2)
)
for y in np.arange(-0.5, image.shape[1], 1.0):
ax.add_artist(Line2D((-0.5, image.shape[1]), (y, y), color='blue', linewidth=2))
if plot_text:
for i, j in np.ndindex(*image_values.shape):
ax.text(
j,
i,
image_values[i, j],
fontsize=8,
horizontalalignment='center',
verticalalignment='center',
color='red',
)
return
def prune(G, node, res):
"""Transform a canonical max tree to a max tree."""
value = G.nodes[node]['value']
res[node] = str(node)
preds = [p for p in G.predecessors(node)]
for p in preds:
if G.nodes[p]['value'] == value:
res[node] += f", {p}"
G.remove_node(p)
else:
prune(G, p, res)
G.nodes[node]['label'] = res[node]
return
def accumulate(G, node, res):
"""Transform a max tree to a component tree."""
total = G.nodes[node]['label']
parents = G.predecessors(node)
for p in parents:
total += ', ' + accumulate(G, p, res)
res[node] = total
return total
def position_nodes_for_max_tree(G, image_rav, root_x=4, delta_x=1.2):
"""Set the position of nodes of a max-tree.
This function helps to visually distinguish between nodes at the same
level of the hierarchy and nodes at different levels.
"""
pos = {}
for node in reversed(list(nx.topological_sort(canonical_max_tree))):
value = G.nodes[node]['value']
if canonical_max_tree.out_degree(node) == 0:
# root
pos[node] = (root_x, value)
in_nodes = [y for y in canonical_max_tree.predecessors(node)]
# place the nodes at the same level
level_nodes = [y for y in filter(lambda x: image_rav[x] == value, in_nodes)]
nb_level_nodes = len(level_nodes) + 1
c = nb_level_nodes // 2
i = -c
if len(level_nodes) < 3:
hy = 0
m = 0
else:
hy = 0.25
m = hy / (c - 1)
for level_node in level_nodes:
if i == 0:
i += 1
if len(level_nodes) < 3:
pos[level_node] = (pos[node][0] + i * 0.6 * delta_x, value)
else:
pos[level_node] = (
pos[node][0] + i * 0.6 * delta_x,
value + m * (2 * np.abs(i) - c - 1),
)
i += 1
# place the nodes at different levels
other_level_nodes = [
y for y in filter(lambda x: image_rav[x] > value, in_nodes)
]
if len(other_level_nodes) == 1:
i = 0
else:
i = -len(other_level_nodes) // 2
for other_level_node in other_level_nodes:
if (len(other_level_nodes) % 2 == 0) and (i == 0):
i += 1
pos[other_level_node] = (
pos[node][0] + i * delta_x,
image_rav[other_level_node],
)
i += 1
return pos
def plot_tree(graph, positions, ax, *, title='', labels=None, font_size=8, text_size=8):
"""Plot max and component trees."""
nx.draw_networkx(
graph,
pos=positions,
ax=ax,
node_size=40,
node_shape='s',
node_color='white',
font_size=font_size,
labels=labels,
)
for v in range(image_rav.min(), image_rav.max() + 1):
ax.hlines(v - 0.5, -3, 10, linestyles='dotted')
ax.text(-3, v - 0.15, f"val: {v}", fontsize=text_size)
ax.hlines(v + 0.5, -3, 10, linestyles='dotted')
ax.set_xlim(-3, 10)
ax.set_title(title)
ax.set_axis_off()
Image Definition#
We define a small test image. For clarity, we choose an example image, where image values cannot be confounded with indices (different range).
Max-tree#
Next, we calculate the max-tree of this image. max-tree of the image
Image plots#
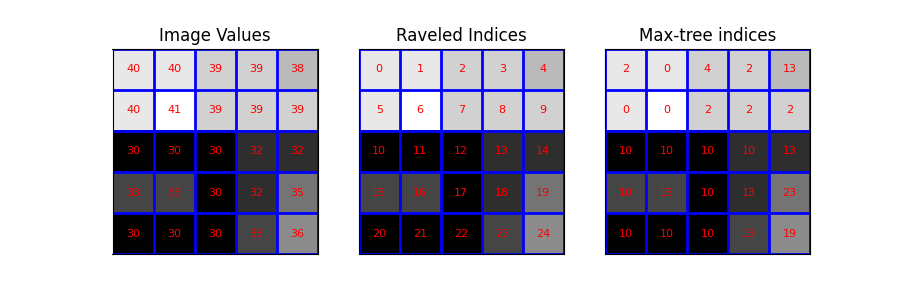
Then, we visualize the image and its raveled indices. Concretely, we plot the image with the following overlays: - the image values - the raveled indices (serve as pixel identifiers) - the output of the max_tree function
# raveled image
image_rav = image.ravel()
# raveled indices of the example image (for display purpose)
raveled_indices = np.arange(image.size).reshape(image.shape)
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, sharey=True, figsize=(9, 3))
plot_img(ax1, image - image.min(), 'Image Values', plot_text=True, image_values=image)
plot_img(
ax2,
image - image.min(),
'Raveled Indices',
plot_text=True,
image_values=raveled_indices,
)
plot_img(ax3, image - image.min(), 'Max-tree indices', plot_text=True, image_values=P)
Visualizing threshold operations#
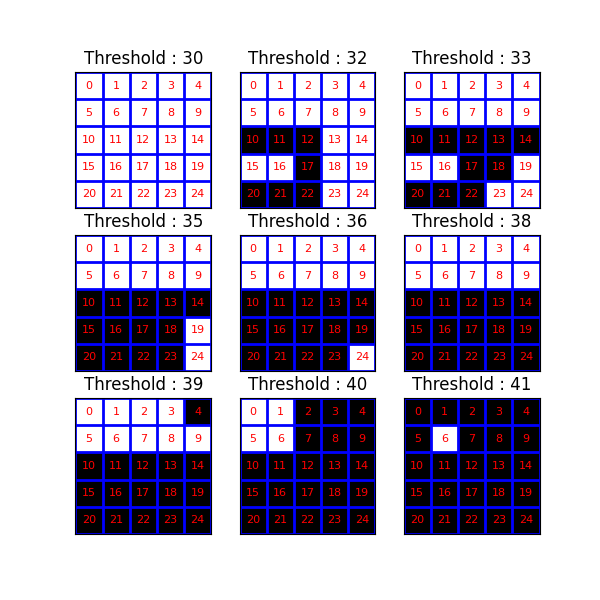
Now, we investigate the results of a series of threshold operations. The component tree (and max-tree) provide representations of the inclusion relationships between connected components at different levels.
fig, axes = plt.subplots(3, 3, sharey=True, sharex=True, figsize=(6, 6))
thresholds = np.unique(image)
for k, threshold in enumerate(thresholds):
bin_img = image >= threshold
plot_img(
axes[(k // 3), (k % 3)],
bin_img,
f"Threshold : {threshold}",
plot_text=True,
image_values=raveled_indices,
)
Max-tree plots#
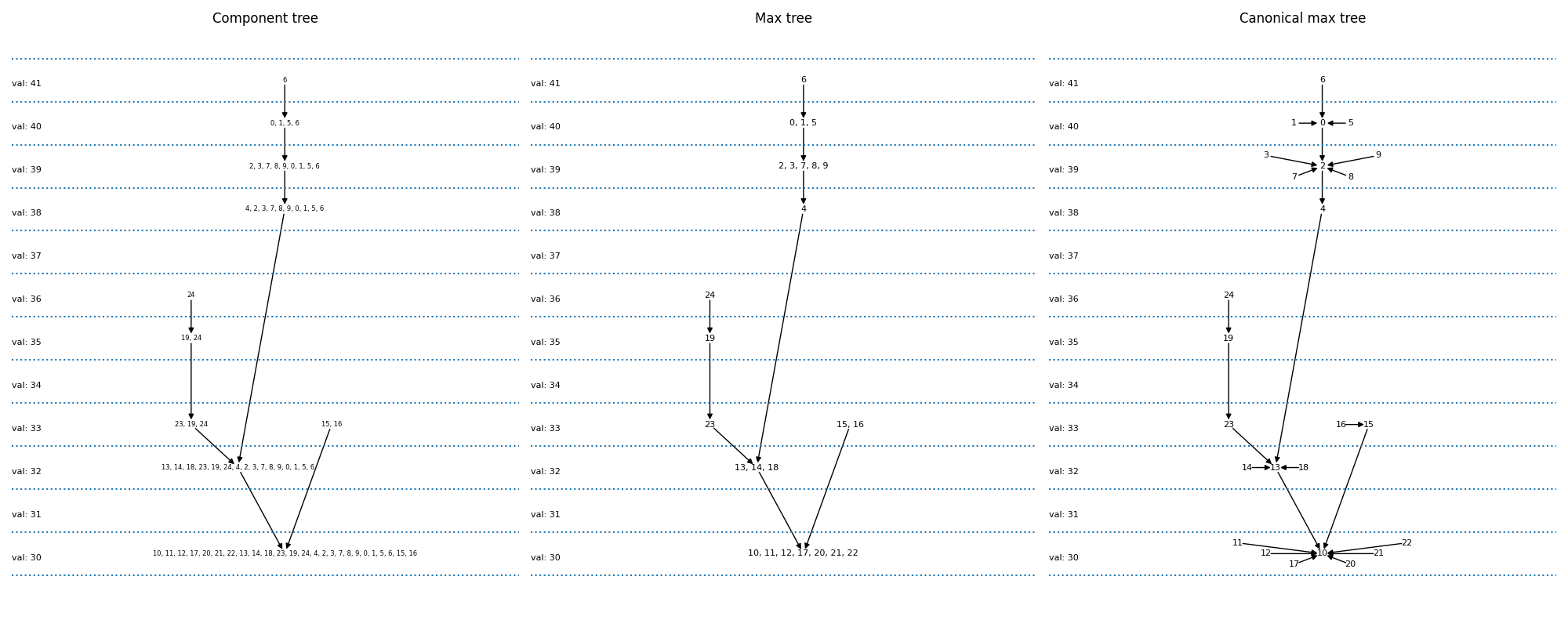
Now, we plot the component and max-trees. A component tree relates the different pixel sets resulting from all possible threshold operations to each other. There is an arrow in the graph, if a component at one level is included in the component of a lower level. The max-tree is just a different encoding of the pixel sets.
the component tree: pixel sets are explicitly written out. We see for instance that {6} (result of applying a threshold at 41) is the parent of {0, 1, 5, 6} (threshold at 40).
the max-tree: only pixels that come into the set at this level are explicitly written out. We therefore will write {6} -> {0,1,5} instead of {6} -> {0, 1, 5, 6}
the canonical max-treeL this is the representation which is given by our implementation. Here, every pixel is a node. Connected components of several pixels are represented by one of the pixels. We thus replace {6} -> {0,1,5} by {6} -> {5}, {1} -> {5}, {0} -> {5} This allows us to represent the graph by an image (top row, third column).
# the canonical max-tree graph
canonical_max_tree = nx.DiGraph()
canonical_max_tree.add_nodes_from(S)
for node in canonical_max_tree.nodes():
canonical_max_tree.nodes[node]['value'] = image_rav[node]
canonical_max_tree.add_edges_from([(n, P_rav[n]) for n in S[1:]])
# max-tree from the canonical max-tree
nx_max_tree = nx.DiGraph(canonical_max_tree)
labels = {}
prune(nx_max_tree, S[0], labels)
# component tree from the max-tree
labels_ct = {}
total = accumulate(nx_max_tree, S[0], labels_ct)
# positions of nodes : canonical max-tree (CMT)
pos_cmt = position_nodes_for_max_tree(canonical_max_tree, image_rav)
# positions of nodes : max-tree (MT)
pos_mt = dict(zip(nx_max_tree.nodes, [pos_cmt[node] for node in nx_max_tree.nodes]))
# plot the trees with networkx and matplotlib
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, sharey=True, figsize=(20, 8))
plot_tree(
nx_max_tree,
pos_mt,
ax1,
title='Component tree',
labels=labels_ct,
font_size=6,
text_size=8,
)
plot_tree(nx_max_tree, pos_mt, ax2, title='Max tree', labels=labels)
plot_tree(canonical_max_tree, pos_cmt, ax3, title='Canonical max tree')
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 1.253 seconds)
