%matplotlib inline
Morphological operations#
Morphology is the study of shapes. In image processing, some simple operations can get you a long way. The first things to learn are erosion and dilation. In erosion, we look at a pixel’s local neighborhood and replace the value of that pixel with the minimum value of that neighborhood. In dilation, we instead choose the maximum.
import numpy as np
from matplotlib import pyplot as plt, cm
import skdemo
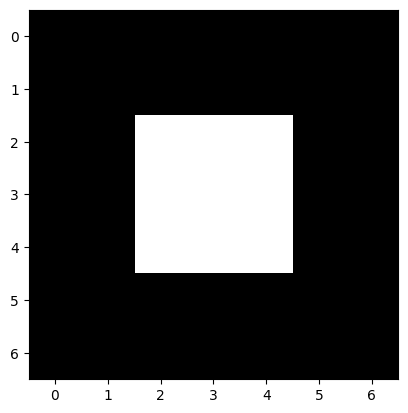
image = np.array([[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0]], dtype=np.uint8)
plt.imshow(image);
The documentation for scikit-image’s morphology module is here.
Importantly, we must use a structuring element, which defines the local
neighborhood of each pixel. To get every neighbor (up, down, left, right, and
diagonals), use morphology.square; to avoid diagonals, use
morphology.diamond:
from skimage import morphology
sq = morphology.square(width=3)
dia = morphology.diamond(radius=1)
print(sq)
print(dia)
[[1 1 1]
[1 1 1]
[1 1 1]]
[[0 1 0]
[1 1 1]
[0 1 0]]
/var/folders/hd/rfxyn9gx4bl39bvwzrgn3rtr0000gn/T/ipykernel_98097/2643900422.py:2: FutureWarning: `square` is deprecated since version 0.25 and will be removed in version 0.27. Use `skimage.morphology.footprint_rectangle` instead.
sq = morphology.square(width=3)
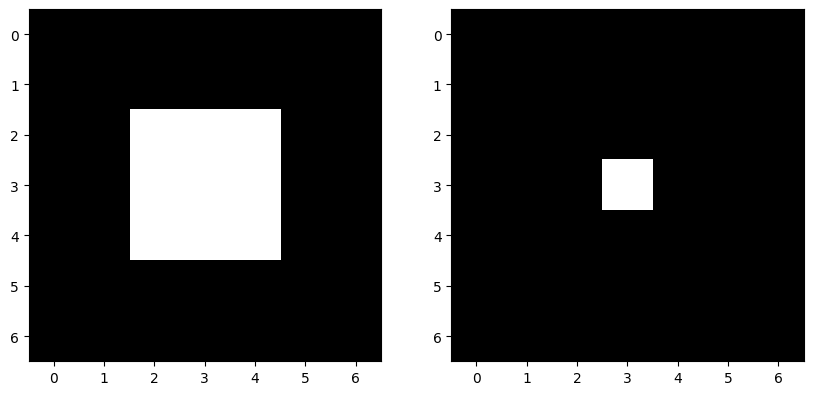
The central value of the structuring element represents the pixel being considered, and the surrounding values are the neighbors: a 1 value means that pixel counts as a neighbor, while a 0 value does not. So:
skdemo.imshow_all(image, morphology.erosion(image, sq))
and
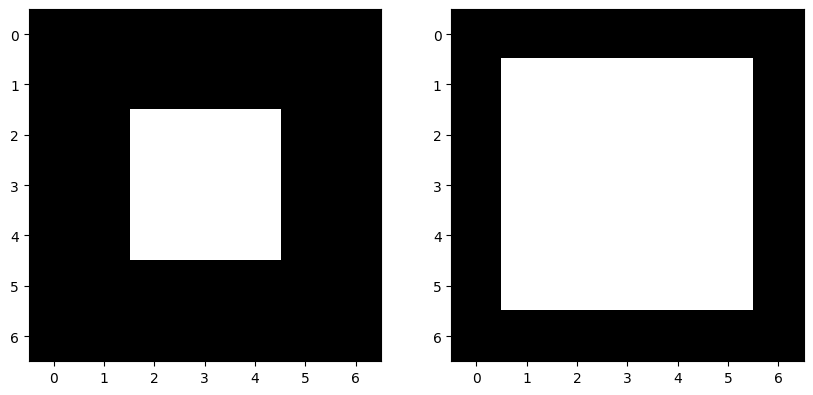
skdemo.imshow_all(image, morphology.dilation(image, sq))
and
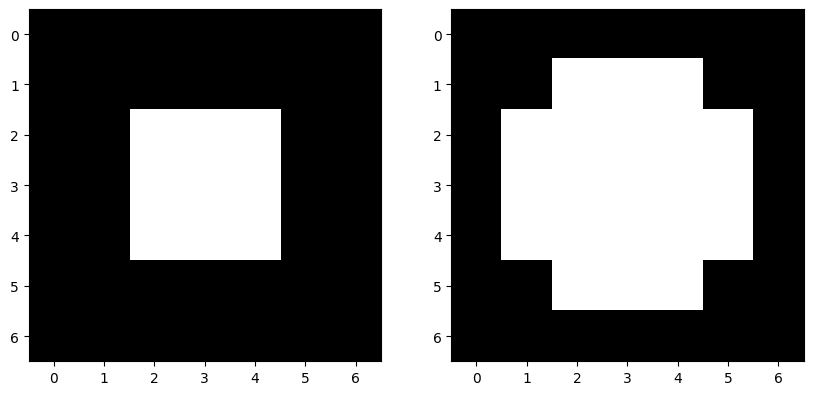
skdemo.imshow_all(image, morphology.dilation(image, dia))
Erosion and dilation can be combined into two slightly more sophisticated operations, opening and closing. Here’s an example:
image = np.array([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], np.uint8)
plt.imshow(image);
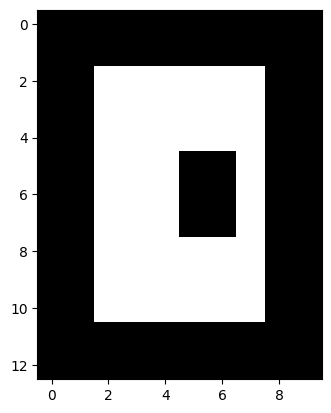
What happens when run an erosion followed by a dilation of this image?
What about the reverse?
Try to imagine the operations in your head before trying them out below.
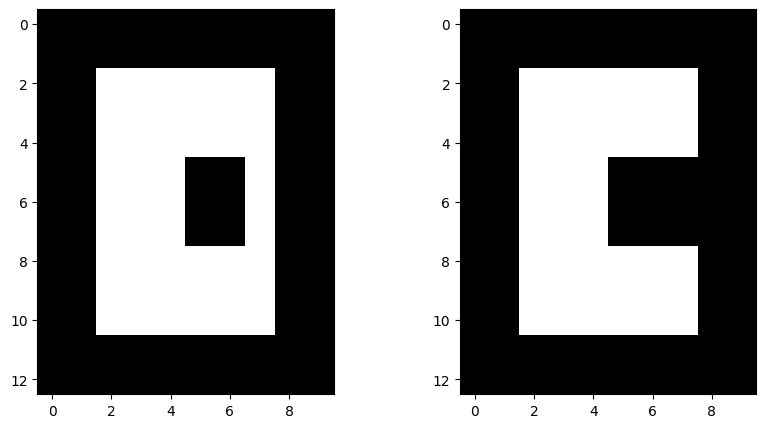
skdemo.imshow_all(image, morphology.opening(image, sq)) # erosion -> dilation
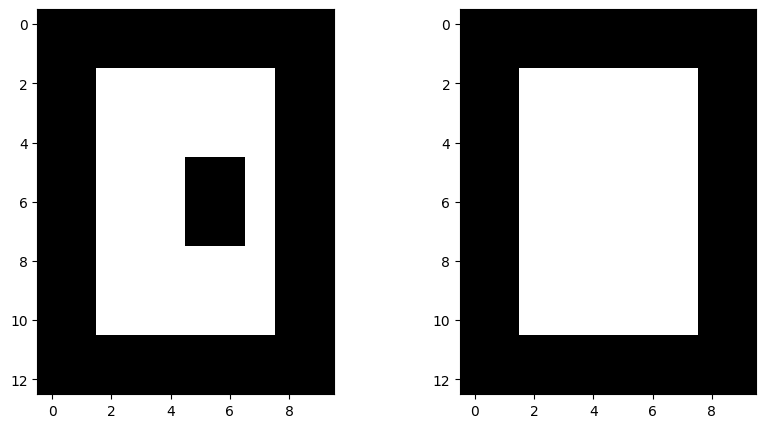
skdemo.imshow_all(image, morphology.closing(image, sq)) # dilation -> erosion
Exercise: use morphological operations to remove noise from a binary image.
from skimage import data, color
hub = color.rgb2gray(data.hubble_deep_field()[350:450, 90:190])
plt.imshow(hub);

Remove the smaller objects to retrieve the large galaxy.
